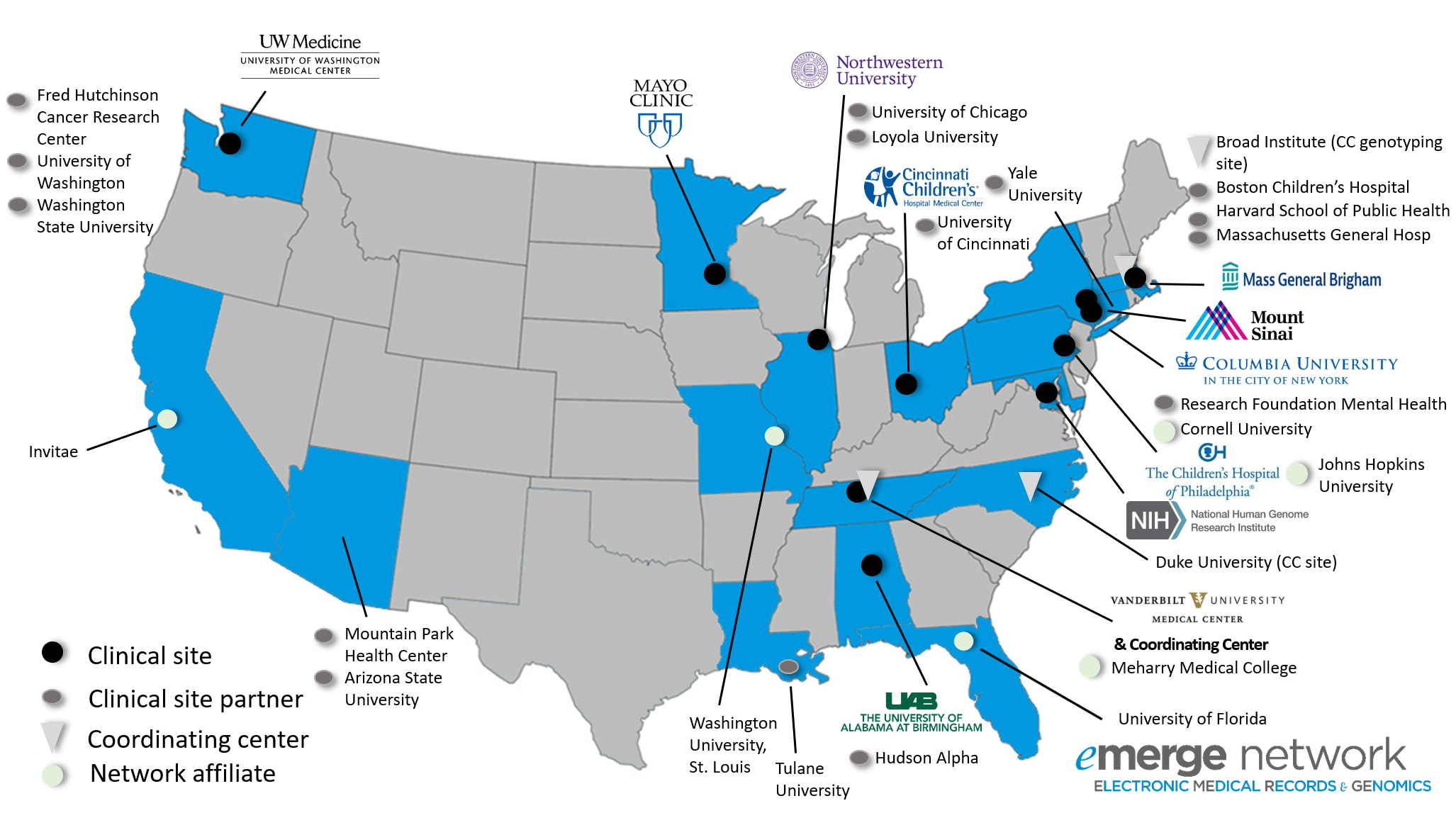
The eMERGE Network brings together researchers with a wide range of expertise in genomics, statistics, ethics, informatics, and clinical medicine from leading medical research institutions across the country. Each center participating in the consortium is uniquely situated to provide critical resources to this highly collaborative and productive network. Each site combines a biobank or study cohort with extensive genomic data and access to clinical data derived from electronic medical records. Sites are geographically dispersed and have diverse patient populations, including two sites focusing specifically on pediatrics. Member sites include:
Participant Sites: Project Overview
Click on each site below for more information:
Children’s Hospital of Philadelphia (CHOP)
Principal Investigator: Hakon Hakonarson, MD, PhD
The Center for Applied Genomics (CAG) is a specialized Center of Emphasis at the Children’s Hospital of Philadelphia (CHOP) with the primary goal of translating basic research findings to medical innovations. The mission of CAG is to develop new and better ways to diagnose and treat children affected by rare and complex medical disorders. We aim to discover genetic causes for the most prevalent diseases of childhood including ADHD, asthma, autism, diabetes, epilepsy, obesity, schizophrenia, pediatric cancer, and a range of rare diseases. Ultimately, our objective is to generate new diagnostic tests and to guide physicians to the most appropriate therapies. Our participants are recruited from the CHOP Health Care Network, which includes over 50 sites throughout Pennsylvania and New Jersey, 100% of which use Epic EMR system. This constitutes the country’s largest pediatric integrated delivery system, supporting >1.1 million visits/year within the primary and specialty care networks, community-based inpatient pediatric and neonatal programs, a rehabilitation hospital, and a home care service. It provides acute- and well-care to patients in urban, rural, and suburban settings.
Cincinnati Children’s Hospital Medical Center (CCHMC)
Principal Investigators: Cindy Prows, MSN, APRN, CNS; Leah Kottyan, PhD; Lisa Martin, PhD
CCHMC has proposed 12 ambitious initiatives for eMERGE IV. The major goals are to generate, return as appropriate along with clinical recommendations, individualized genomic risk assessments that include polygenic risk scores for 2500 minority participants ascertained on dyads (mother/newborn) and extended to families (dyad + biologic father, siblings). The informatics initiatives include integration of electronic medical records and development, implementation and evaluation of clinical decision support at two different hospital systems, one pediatric and one adult. Participant preferences, education needs, and ethical and social implications will be studied throughout the 5 year period. A unique contribution to the Network will include CCHMC funded low read depth coverage (LRDC) whole genome sequencing data for at least 30,000 individuals along with phenotyping data for further discovery research and comparison of PRS derived from LRDC and commercially available genotyping arrays for Network selected phenotypes.
Columbia University
Principal Investigators: Wendy Chung, PhD, MS; George Hripcsak, MD, MS; Krzysztof Kiryluk, MD; Chunhua Weng PhD, MS
Columbia University in partnership with the New York-Presbyterian hospital has a long and strong tradition of patient-centered research and community engagement by serving a socioeconomically, racially, and ethnically diverse community in the urban, medically underserved communities of Washington Heights and Harlem in Northern Manhattan, and has established Precision Medicine as a major university-wide initiative (PMI), of which eMERGE is a vital program. We possess broad and deep expertise in clinical genetics, genomics, informatics, EHR phenotyping, systems biology, public health, and ELSI, including return of genetic results and secondary findings to diverse patients. Columbia has also made significant contributions to the goals of eMERGE III project, including leading the effort to leverage the OMOP Common Data Model (CDM) to improve the efficiency and accuracy of using heterogeneous EHR data for developing reproducible and portable electronic phenotype algorithms; contributing a widely-adopted XML parser for structuring genomic test reports; contributing to the development of a shared dataflow pipeline for eMERGE III; understanding the diagnostic limitations in sequencing technologies and their implications for genomic health disparities; assessing the cost-effectiveness of different recruitment methods in eMERGE III; sharing lessons learned in linking EHR and sequence data for genomic decision support, developing scalable genomic education methods and assessing the impact of returning genetic results on patients, families, and clinicians. We are poised to contribute novel comprehensive risk prediction and management solutions for diverse patients as part of eMERGE IV efforts.
Icahn School of Medicine at Mt. Sinai (Mt. Sinai)
Principal Investigator: Eimear Kenny, PhD; Noura Abul-Husn, MD, PhD
The Mount Sinai Health System serves some of the most diverse patients in the world, and Mt. Sinai investigators have been at the center of many NHGRI-led genomic research (GSP, PAGE) and genomic medicine (eMERGE, CSER, IGNITE) consortia. Building on our decade-long track record in genomic research, clinical trials, and genomic medicine in diverse, underserved patients from New York City, we propose to develop new frameworks to bring genomic risk into clinical care to promote health equity. In addition to genomic discovery and risk prediction, we are creating analytical and data resources, and digital health applications tailored to diverse populations, as part of the eMERGE IV Network. The Mt. Sinai team is composed of clinicians and scientists who work closely with community members and industry partners to develop flexible and innovative strategies to bring leading-edge research into clinical care. Having recently announced a large institutional investment to scale the integration of genomics and artificial intelligence into patient care, Mt. Sinai is poised to contribute as an enhanced diversity clinical site to eMERGE IV’s mission to implement and learn from genomic risk assessment in clinical care. Mt. Sinai’s team will contribute to progress in multiple areas across eMERGE, including developing and deploying electronic phenotypes; discovering new genotype-phenotype associations; and facilitating the communication of genomic results to ancestrally and socioeconomically diverse patients and their healthcare providers.
Mass General Brigham
Principal Investigators: Elizabeth Karlson, MD; Shawn Murphy, MD, PhD; Jordan Smoller, MD, ScD
The eMERGE IV Clinical Center at Mass General Brigham leverages a large biobank, clinical data in the electronic health records (EHR) for >6 million patients from the largest integrated health care provider in New England, advanced bioinformatics expertise, prior leadership in PRS development, established expertise in returning genomics results, and experience using information technology to transform clinical processes and assess medical, behavioral and economic outcomes. We have developed highly specific machine learning algorithms for e-phenotypes and implemented eMERGE algorithms for multiple phenotypes and have considerable experience in developing decision support tools and analyzing outcomes that can be leveraged to study the integration of PRS data into clinical practice. We propose to build on our expertise to accomplish the following specific aimsAim 1, Discovery. We will test the hypothesis that PRS will allow us to stratify eMERGE participants based on genetic risk for common complex traits. We will develop new PRS for coronary artery disease, atrial fibrillation, type 2 diabetes, colorectal cancer and depression in diverse GWAS populations and validate multi-ethnic PRS in the Mass General Brigham Biobank and eMERGE III dataset. We will obtain PRS from our network colleagues for an additional 15 electronic phenotypes with a goal of identifying high-risk subjects Aim 2. Report development. We will develop a “Risk Insight Report” combining clinical risk factors, family history, and PRS with evidence-based recommendations for high risk participants ( e.g. top 2% of PRS distribution) for electronic clinical implementation. We will analyze the independent contribution of clinical risk factors, family history, and PRS using multivariable methods. Our ELSI subaim will test the impact and interpretability mock Risk Insight Reports. We will assess the extent to which the mock reports are understood by both providers and patients to minimize harms and maximize benefits of genomic risk assessment in eMERGE IV; Aim 3. Implementation. We will test the hypothesis that health-care providers will alter their surveillance and treatment of participants based on electronic clinical decision support (eCDS) of Risk Insight Reports. Among providers for high-risk participants, we will see at least one change in clinical care after disclosure discussions with participants (eg. assess ordering screening tests, prescribing a preventive therapy, or providing lifestyle advice). We will conduct an analysis of the effect of disclosing results to high risk participants to determine how personalized results changed patient outcomes in laboratory values, risk reduction behaviors, or health care utilization.
Mayo Clinic (Mayo)
Principal Investigators: Richard Sharp, PhD
The goal of Mayo eMERGE investigators is to leverage the electronic health record (EHR) for genomic discovery and to implement genomic medicine to improve health care outcomes, working closely with other eMERGE sites and the Mayo Center for Individualized Medicine. In eMERGE I we used genotype data and electronic phenotyping algorithms for genomic discovery. In eMERGE II we conducted a clinical trial of disclosing a polygenic risk score (PRS) for coronary heart disease (CHD) and placed pharmacogenomic variants in the EHR with linkage to clinical decision support (CDS). In eMERGE III, we returned actionable findings from targeted sequencing of 68 genes and collaborated with a Federally Qualified Health Center (FQHC) to bring genomic medicine to an under-represented population. In eMERGE IV, as part of the Genomic Risk Assessment & Management Network, we will test the hypothesis that individuals at high polygenic risk for ≥1 of 15 common diseases will have a higher uptake of risk reduction measures than those at lower risk. We will build on our prior work in genomic discovery, EHR integration and return of results (RoR) to develop and validate PRS for 15 diseases and combine these with clinical risk scores and family history to compute comprehensive risk profiles for 2500 individuals (35% non-European ancestry). Our prior work in disclosing a PRS for CHD to research participants as part of a comprehensive risk profile enables us to efficiently expand the approach to other common diseases that pose a significant public health burden. To assess the utility of PRSs, we will recruit 1800 adults and 200 adolescents from Rochester MN and 500 adults from Phoenix AZ. DNA will be sent for CLIA-certified genomic testing and we will calculate ancestry specific PRSs for each participant. Individualized risk estimates will be reported to participants and health care providers (HCPs) and placed in the EHR with linkage to CDS that includes guideline-based risk-management information. For participants who are at significantly higher polygenic risk of developing at least 1 of the 15 diseases, results will be disclosed in-person by a HCP followed by assessment of outcomes including new tests ordered, risk reduction measures, and changes in modifiable risk factors. We will examine the economic, behavioral and psychosocial implications of returning PRS. Our focus on PRS for common cardiovascular diseases brings genomic medicine to the population. We will continue our partnership with Mountain Park Health Center, a FQHC serving low-income, minority communities in Phoenix AZ, to address concerns about health disparities in the context of genomic medicine.
Northwestern University (NU)
Principal Investigators: Rex Chisholm, PhD; Elizabeth McNally, MD, PhD; Laura Rasmussen-Torvik, PhD
Northwestern University is dedicated to translational genomics, with a strong focus on unified, interdisciplinary research bridging the informatics, clinical, basic science, and bioethics domains. Over the phases of eMERGE, Northwestern has applied this interdisciplinary approach to study the feasibility, utility, outcomes, and effectiveness of EHR-linked genomic information as a member of the network. Particular areas of focus include: novel informatics approaches to integrate genomic results into EHR systems; development of open-source, EHR-based phenotyping algorithms to empower translational research across a variety of clinical domains; and studies of participant, patient, and provider interaction with EHR-based genomic results. In phase IV of eMERGE, Northwestern is building on this expertise to develop, deploy, and evaluate systems of EHR-based, genomics-guided clinical risk assessment and management tools across a variety of common, impactful conditions. Northwestern’s cross-disciplinary approach to this work is empowered by an innovative smartphone app to consolidate outreach, enrollment, engagement, return of results, and dissemination of health education materials surrounding genetic risk and risk management. Focusing recruitment on community-based clinics, Northwestern aims to engage participants from communities that, though not traditionally included in academic research settings, could benefit significantly from these risk management approaches. By combining expertise in informatics and EHR integration with innovative return-of-results strategies rooted in behavior theory, Northwestern aims to provide valuable data on accuracy, utility, and uptake of genetic risk assessment as the field moves towards integrating genetic risk scores into clinical practice.
University of Alabama at Birmingham (UAB)
Principal Investigators: Nita Limdi, Pharm.D, PhD, MSPH, FAHA; James Cimino, PhD
Our eMERGE team aims to bring our experience and expertise in research with medically underserved (>75%) and African American (30-40%) populations to identify genomic signatures of common diseases, conduct genomic risk assessments within the clinical workflow and where appropriate, pre-treat at-risk patients with common diseases including diabetes, chronic obstructive pulmonary disease, obesity, coronary artery disease and prostate cancer. Our aims will also allow us to develop genomic risk assessments for African Americans, and address challenges involved in bringing genomic medicine into routine medical care.
Working with the eMERGE network, we will select the fifteen diseases of public health impact and select polygenic risk scores, (making adaptations (if any) for African Americans) for clinical implementation. To strengthen the evaluation of race-specific genomic risk, we bring an additional cohort of over 25,000 AAs.
We will conduct an ethical legal social implications (ELSI) study, to explore patient perspectives on the use of family history and polygenic risk scores for estimating disease risk among Alabama Genomic Health Initiative (a statewide cohort) participants. The results will inform the development of consent, educational materials, and a communication strategy to enhance recruitment and retention of eMERGE participants.
Recruiting over 75% of patients from medically underserved communities, we will incorporate genomic risk, family history, and clinical data to identify high-risk patients, recommending risk reduction recommendations in concordance with practice guidelines. For high-risk patients, we will deploy clinical decision support and present the genomic risk assessments and risk reduction recommendations. We will assess three outcomes: uptake of recommendations (implementation outcome), adherence to clinic visits (engagement outcome), and surrogates of disease/control (clinical outcome; e.g. blood sugar, cholesterol). We bring our expertise and experience to collaborate with the eMERGE investigative team to leverage the power of genomics to prevent disease.
University of Washington (UW)
Principal Investigators: Gail Jarvik, MD, PhD; David Crosslin, PhD
The eMERGE IV Northwest partnership includes clinicians, scientists, and professionals in other fields from University of Washington (UW) Medicine, UW Health Sciences, the Fred Hutchinson Cancer Research Center, and the Brotman Baty Institute for Precision Medicine. The eMERGE IV Northwest partnership aims to evaluate the use of genomic information in the health care of diverse participants at the University of UW. Taking advantage of the extensive UW longitudinal electronic health record (EHR) data linked to the Brotman Baty Institute Biorepository, enrollment prioritizes diversity of ancestry and sexual and gender minorities (LGBTQ) groups that may be under-represented in medical research. Our site is expected to lead work on the prediction of colon cancer, glaucoma, and cardiovascular disease; other conditions expected to be studied include diabetes, liver disease, high cholesterol, and osteoporosis. Working with our eMERGE colleagues, we will aggregate, into a single score that informs about the risk of a disease, a participant’s polygenic test information, which looks at variants from across the genome, as well as other known risk factors, such as personal and family medical history, environmental and social determinants, and physical and lab results. This score would let clinicians know which patients might benefit from screenings or other interventions to reduce the risk of disease or make an early diagnosis. In addition to experts specializing in each of the medical conditions that are the focus of the Northwest site, leaders in health economics, and in the ethical and societal aspects of genomics and health care will also be part of the research team. Medical, as well as economic, outcomes will be measured. Stakeholder engagement will be encouraged to obtain comments from participants and their families.
Vanderbilt University Medical Center (VUMC)
Principal Investigators: Dan Roden, MD; Digna Velez Edwards PhD, MS; Wei-Qi Wei MD, PhD
Vanderbilt University Medical Center has embraced Personalized Health and Healthcare as a key pillar of our Research Strategic Plan. This institutional commitment has taken multiple forms including the development of key infrastructure resources (the institutional Biobank (BioVU); the program in preemptive pharmacogenetic testing; the genotyping and sequencing core) and support of faculty in key disciplines such as genomics, informatics, ethics, privacy science, and clinical pharmacology. VUMC’s team has contributed importantly to progress in multiple areas across eMERGE, including developing and deploying phenotype algorithms; developing and managing the genotype quality control pipeline for the network (eMERGE I); discovering new genotype-phenotype relations in VUMC and cross-network datasets; actively participating in the PGx clinical implementation project, community consultation, and return of results initiatives; developing methods to resolve tensions between data access and individual privacy; developing sampling methodology for the CERC survey; and developing new software tools for the field, including for developing and validating phenotypes, de-identification and for the phenome-wide association study paradigm (“PheWAS”)
eMERGE Network Coordinating Center (CC)
Principal Investigators: Josh Peterson, MD, MPH (VUMC); Niall Lennon, PhD (Broad)
Vanderbilt University Medical Center has been the coordinating center of the eMERGE Network since inception. During this phase of eMERGE we have partnered with the Broad Institute to to add expertise in large-scale genotyping, cloud computing, and genomic data analysis and management, and with Duke University to leverage its proficiency in the implementation of patient-oriented family history tools and delivery of risk assessments. To meet the needs of the Network, the CC continues to serve NHGRI, the Network, and its members in all coordination, communication, and collaboration functions while providing a nationally visible home for eMERGE authored tools and scientific productivity. The CC is also assisting the Network with selecting and validating polygenic risk scores (PRS) for approximately fifteen complex diseases and developing genomic risk assessments (GRAs) which combine the PRS with other clinical risks, family history, and monogenic risks to individuals of diverse ancestry. Finally, The CC will coordinate a prospective study for 25,000 participants which calculates clinical and genomic risks for participants and their providers. In addition to providing the genotyping and PRS reporting services, we will develop with the Network scalable EHR-based methods to assess uptake of risk-reduction recommendations and clinical impact.
External Scientific Panel
ESP Background
The Network receives regular guidance from a panel of experts, external to the Network member sites. The External Scientific Panel (ESP) holds annual conference calls with the principal investigators from each site, and the ESP additionally attends one in-person meeting each calendar year. Recommendations from the ESP are gathered and addressed by the PI Group and disseminated through the Network for response and action by each.
Click on the member below for more information:
Members