Phenotyping: Cohort discovery using EHR data
Return to Projects Page.
Phenotyping is the practice of developing algorithms designed to identify specific phenomic traits within an individual. These algorithms are created using multiple variables, thus enabling researchers to accurately identify traits and perform analyses. Best practice materials and data standardization tools have been developed to aid with phenotyping protocols and collaboration. The Phenotyping Workgroup also seeks to advance the science of de-identification, transportable phenotyping methods, structure and standards, and portable components of algorithms and methods. Within the eMERGE Network, computable phenotype algorithm development focuses both disease-related and pharmacogenomic-related phenotypes.
Creating Phenotypes
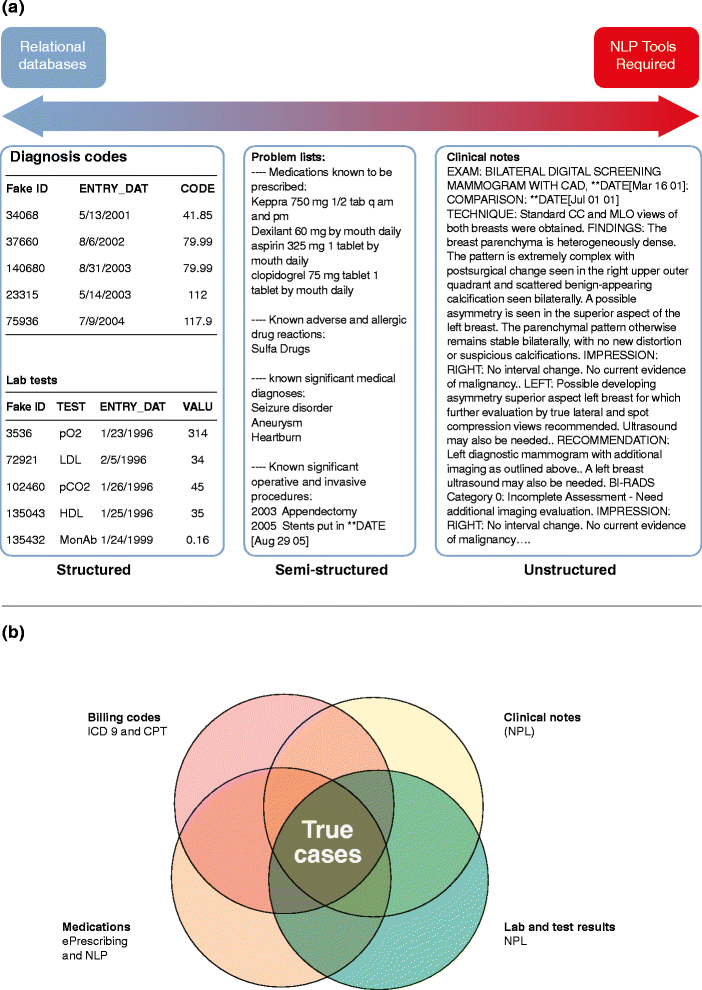
A variety of data can be extracted from electronic health records including structured and unstructred formats, billing codes, laboratory results, medication data and natural language processing (NLP) which searches text (like doctors notes and reports) for key words and information. The ability to pull the correct patient records from a phenotype depends on narrowing down the proper search criteria to target the ‘true’ case. See figure below and learn more, here.
EHR data structure and accurate phenotyping. (a) Electronic health record (EHR) data can be structured or unstructured. Structured data are easy to retrieve whereas unstructured data require additional tools to be used for phenotyping, such as natural language processing (NLP). (b) Accurate phenotyping often requires extracting information from billing codes, prescriptions, laboratory tests and clinical notes. This information can be either structured or unstructured. ICD-9, International Classification of Diseases, Ninth Revision.
Figure and caption courtesy of: Wei-Qi, W. & Denny, J.C. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Medicine, 2015 April 30; 7:41. PMID: 25937834
Phenotype Algorithms
Current phenotyping efforts span multiple diseases in both pediatric and adult participants including adult familial hypercholesterolemia, colorectal cancer, migranes, epilepsy, chronic rhinosinusitis, chronic kidney disease, and hearing loss, with many more currently in development.
* Click here to view a listing of published eMERGE Phenotypes on PheKB.
Phenotype Data
* Click here to view a listing of eMERGE Studies currently available in dbGaP.
* Click here to view a listing of phenotype data collected for eMERGE studies.
Common Variables collected across datasets
- Demographics: sex, decade of birth, year of birth, race & ethnicity
- Codes (repeated values & age): ICD, CPT
- BMI: (repeated value & age): BMI, height, weight
- Labs (name, repeated value & age): Serum total cholesterol, LDL, HDL, Triglycerides, Glucose fasting/nonfasting, autoimmune, & White Blood Cell count
- Meds (repeated name/rxcode & age): Cerivastatin sodium, Rosuvastatin, Simvastatin, Fluvastatin, Pravastatin, Lovastatin, Atorvastatin, & Pitavastatin
Tools
eRC | eMERGE Record Counter, provides exploratory data figures for research planning purposes and feasibility assessment
PheKB | Phenotype Knowledgebase, offers a collaborative environment to build and validate electronic phenotype algorithms
PheWAS | PheWAS Catalog, functions as a platform for analysis of phenotypes against single gene variants
SPHINX | Sequence and Phenotype Integration Exchange, tool for exploring data for hypothesis generation, especially around drug response implications of genetic variation across the eMERGE PGx cohort