The eMERGEseq platform
Return to Projects Page.
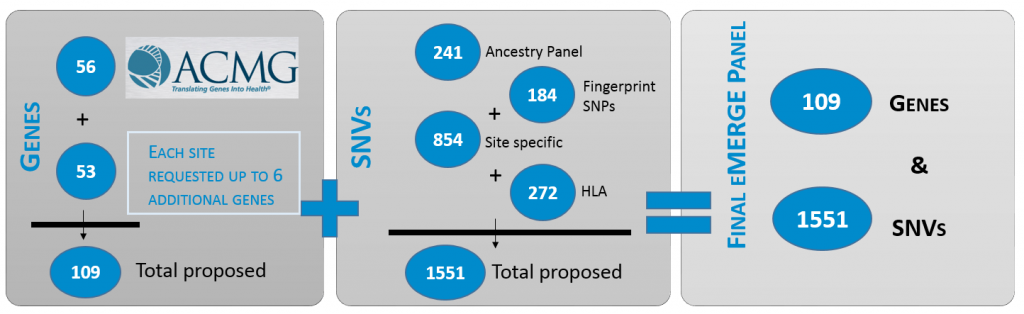
One of the major aims of the eMERGE Network during Phase III is to identify rare variants with presumed major impact on function in a cohort of 25,000 participants across the Network. The Network created an eMERGE specific sequencing platform that is used to sequence participants at the individual sites. Baylor College of Medicine Human Genome Sequencing Center (HGSC) and Partners Healthcare with Broad Institute (the two sequencing centers) worked with the Clinical Annotation Workgroup as well as the individual eMERGE sites to identify and validate an impactful set of genes and single nucleotide variants (SNVs) that allow for clinically actionable, pathogenic variants to be returned while providing researchers with the data needed to aid in genomic discovery. This resulted in a panel consisting of 109 genes and 1,551 SNVs.
Creating the panel
Determining which genes to return to participants
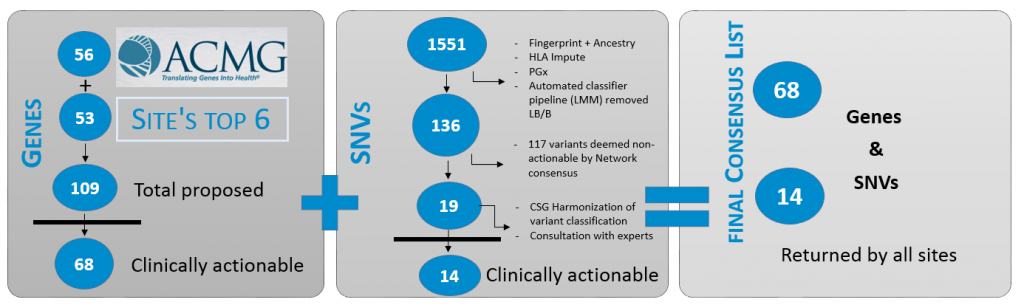
The Network has a dedicated workgroup, Clinical Annotation, that focuses on building consistency of approaches to the gene and variant interpretation across the eMERGE sequencing centers and study sites as well as supporting contribution to public knowledge base, including submissions to ClinVar. This workgroup analyzed the genes and SNVs on the eMERGEseq panel assess clinical validity and actionability to determine which genes and SNVs should be returned to participants. The Clinical Annotation workgroup determined that of the 109 genes and 1,551 SNVs on the panel, 68 genes and 14 SNVs warranted return to participants. This group also reviews questions that arise during sequencing and case review including challenging variants as well as any variant interpretation descrepancies that are not resolvable between the CSGs to maintain consistency throughout the network. View the list of actionable genes and SNVs here.
Disease categories
The 68 returnable genes have variants that have been implicated in the following disease processes: Cancer susceptibility & tumor disease, cardiac disease, hypercholesterolemia, diabetes & kidney disease, Ehlers-Danlos Syndrome, neuromuscular disease, and Ornithine Transcarbamylase (OTC) deficiency. The 14 returnable SNVs have been related to the following disease categories: inborn error of metabolism, cancer susceptibility, endocrinology, clotting disorders, iron storage, and inflammatory diseases.
Sample collection and processing
All blood samples are collected from consented participants and DNA is extracted at the individual sites. The samples are barcoded and shipped to the Sequencing Centers where they undergo quality control testing and processing. Samples are maintained in a CLIA compliant manner throughout the whole collection and sequencing process, allowing results to be returned to participants. Clinical results on the 68 actionable genes and 14 SNVs are returned to sites in both a PDF and structured data format, allowing for integration into the electronic health record. Research data on the remainder of the genes and SNVs are also returned to the sites. Click on the hyperlink to learn more about the Return of Results and EHR integration process.
Participants per site
Tools
- ACMG recommendations: The American College of Medical Genetics and Genomics has published recommendations for reporting incidental findings in the exons of certain genes
- SNV: Single Nucleotide Variants are very similar to Single Nucleotide Polymorphisms (SNPs), which are the most common type of genetic variation among people. This is when one nucleotide in a persons DNA is different compared to the rest of the population. However SNVs are not required to be at a certain frequency or threshold in the population. A SNV can be unique to one individual, where many people in a population are likely to have the same SNPs.
- CLIA compliance: Learn more about requirements and standards needed for return of genetic results to participants.