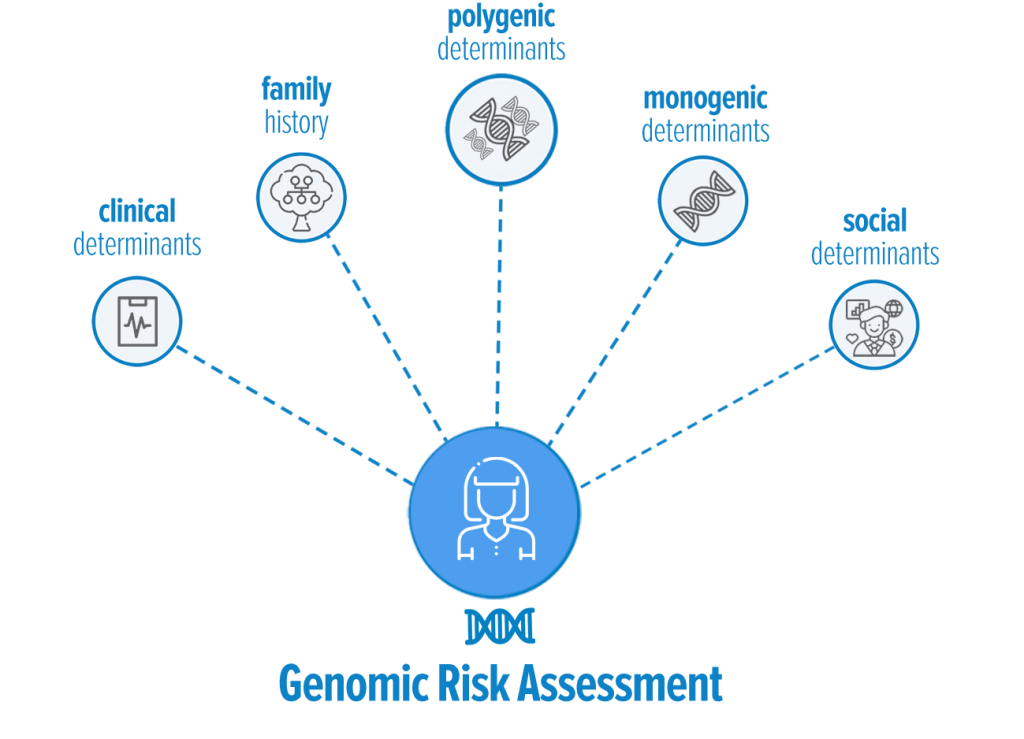
Genomic Risk
A major goal of the eMERGE network is to advance genomic research to allow doctors to better predict if and when a person will develop a condition. Determining why some individuals are prone to diseases like cancer or cardiac conditions can help physicians identify risk factors early, potentially allowing for earlier screening, diagnostics, or interventions, than conventionally available. Development of a disease is complex, and can have many factors. Some diseases are strongly influenced by one or a few genes, others by multiple genes, a persons environment, health history, family history, and access to health care. Integrating how all these different risk factors contribute to development of a disease is a major focus of genomic research. Below are some details and links to explain how these different risk factors influences a persons overall risk of developing a given condition.
Monogenic determinants
Monogenic diseases are a result of a change in a single gene, they are inherited according to Mendel’s Laws and sometimes called Mendelian disorders. Some commonly know monogenic disease are conditions like Cystic Fibrosis and Sickle Cell Anemia. However, monogenic changes can also contribute to risk of other types of conditions like breast cancer. Development of breast cancer has many contributors, but in some cases Mendelian inheritance of a gene called BCRA 1/2 can increase an individuals risk. During a previous round of the eMERGE network (more information here), we examined how monogenic pathogenic variants in genes associated with conditions like Hereditary Breast and Ovarian Cancer, Lynch Syndrome, and Familial Hypercholesterolemia, know as Tier 1 conditions. The network is currently determining how risk of monogenic disease integrates with other risk factors like polygenic and family history to determine how overall genomic risk contributes to disease development and progression.

Polygenic determinants
Polygenic determinants and is when many genes or variations in gene across your genome contribute to the development of a disease. By examining how the total number of changes across the genome are associated with development of disease allows scientists to understand the complexity of disease development across populations. Learn more about polygenic risk on the NHGRI website and the Broad Institute.

Family History
Family history can play an important risk in the development of a disease. Families can share genes, environments, and lifestyle. In order to understand total genomic risk, it is important to take into account family health history. Learn more about why it is important to study family health history on the NIH website. The eMERGE Network is utilizing the MeTree tool, developed by Duke University, to capture family health history and integrate with other risk factors to examine overall genomic risk.

Clinical determinants
Clinical risk factors are physiological factors that at certain levels contribute to the development of a disease. These factors are typically measured clinically, by a doctor or health care professional, and can include things like laboratory results, weight or BMI, physiological tests like blood pressure, or previous health history. By taking into consideration a person’s clinical risk factors these data can be examined in combination with polygenic determinants, monogenic determinants, and social determinants of health to better understand a person’s overall genomic risk of developing a given condition.

Social Determinants
Social determinants of health (SDOH) are how the environment a person lives in, grows up in, works in, and interacts with that can have influences on their health and outcomes of diseases. Five major SDOH include Economic stability, Education access & quality, Health care access & quality, Neighborhood & environment, and Social & community interactions. Learn more about SDOH on the Healthy.gov and World Health Organization websites.

