Genomics
Return to Projects Page.
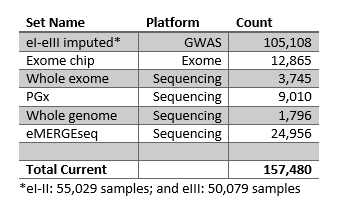
Genotyping is a process in which an individual’s DNA is examined to reveal specific traits and alleles. The eMERGE Network has undertaken many genotyping projects since the foundation in 2007. The following describe the data sets created by the eMERGE Network. The Genomics Workgroup guides data QC and analysis throughout the Network. A listing of the eMERGE studies currently available on dbGaP can be found here. A list of phenotype data available can be found here. Please see the below tables for a break down of count by site and platform. General descriptions of the datasets can also be found on the NHGRI eMERGE webpage.
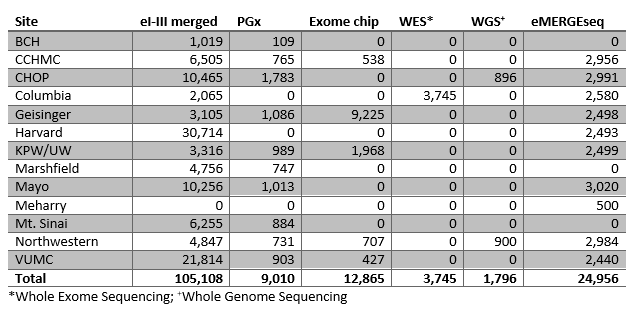
eMERGE I-III imputed data set (N = 105,108)
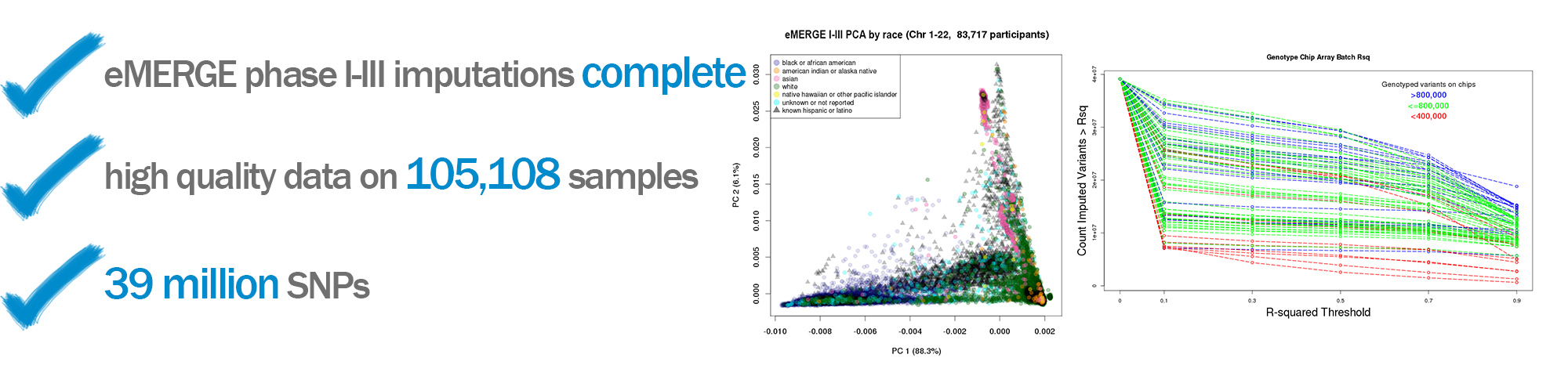
The eMERGE Network also uses data from participants that are genotyped by individual sites, and this individual level data is then combined to create a de-identified dataset used for analysis across the Network. All data are sent through a strict quality control protocol and imputed to guarantee consistency and quality. Recently the eMERGE I-III GWAS data were imputed off the Haplotype Reference Consortium (HRC reference) using the Michigan Imputation Server. This produced a dataset with over 105,000 samples that is linked to phenotypic information across the sites. Using 1000 Genome structural variants an imputed array of the same 105,108 data sets was also produced during Phase III.
eMERGE exome chip data set (N = 12,865)
Five sites across the Network provided exome chip data at the beginning of Phase III. The exome chip genotype content is based on the deep sequencing of ~12,000 African Ancestry and European ancestry individuals to discover rare variants which are then typed on this chip. The exome chip genotype also contains many of the variants associated with disease in the GWAS Catalog and a selection of common variants useful in controlling for genetic ancestry in association studies.
eMERGE PGx data set (N = 9,010)
eMERGE-PGx was a multi-site test of the concept that genetic sequence information can be coupled to electronic medical records (EMRs) for use in healthcare. This project focused on pharmacogenomics (PGx) – the idea that individual variation in drug response includes a genomic component. The Network used the PGRNSeq sequencing platform developed by the PGRN Network. PGRNSeq was developed in a cooperation between Deep Sequencing Resources at the Baylor College of Medicine and the University of Washington to fully characterize the genetic variation across human populations in 84 genes. Though this project completed during Phase II of the eMERGE Network, work is still ongoing. Sequencing and phenotype data are housed in SPHINX (Sequence, PHenotype Integration Exchange).
eMERGEseq data set (N = 24,956)
During this phase the Network has launched an effort to sequence DNA from 25,000 participants across the consortium. These genotypes were sequenced on an eMERGE specific panel based off of ACMG 56 and the results have been returned to the participants. Learn more about the eMERGEseq panel here.
eMERGE Whole Genome Sequencing data set (N = 1,796)
Nine hundred Whole Genome Sequencing (WGS) data are available from each CHOP (pediatric subjects) and Northwestern (adult subjects). These subjects are racially and ethnically diverse and include European, African and Hispanic ancestry. All subjects also have electronic health record information at their respective site, allowing for potential genotype/phenotype analysis.
eMERGE Whole Exome Sequencing data set (N = 3,745)
Whole Exome Sequencing (WES) data are available from Columbia. This cohort includes patients identified from our Breast Cancer Family Registry (15%), Pulmonary Artery Hypertension cohort (2%), and the Pediatric Cardiac Genomics Consortium (83%).
Data counts by site