Phenotyping Tools
One of the missions of the eMERGE Network is to share resulting best practices, expertise, and experience within and outside eMERGE and disseminate association findings, tools and best practices to the scientific community. You will find additional resources related to our phenotyping tools.
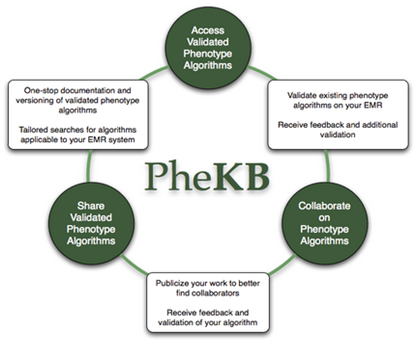
PheKB
The Phenotype Knowledgebase website, PheKB, is a collaborative environment to building and validating electronic phenotype algorithms. The PheKB has both a public component that allows you to view existing phenotypes and implementations, and a collaborative component that allows you to submit phenotypes, provide detail about implementations of a phenotype or add to existing phenotypes. For the collaborative component, you will need to register with the site.
PheKB Presentation (PPT)
PheWAS
Phenome-wide association studies (PheWAS) analyze many phenotypes compared to a single genetic variant (or other attribute). This method was originally described using electronic medical record (EMR) data from EMR-linked in the Vanderbilt DNA biobank, BioVU, but can also be applied to other richly phenotyped sets. Download PheWAS code translation table and Perl Code. Download PheWAS R package. Synthesis-View, PheWAS-View and Phenogram provides visualization tools for genome and phenome-wide data. These tools are also offered via a web interface at http://visualization.ritchielab.psu.edu/.
OMOP
The eMERGE sites have translated their eMERGE cohorts into the OMOP Common Data Model, v5.2.

